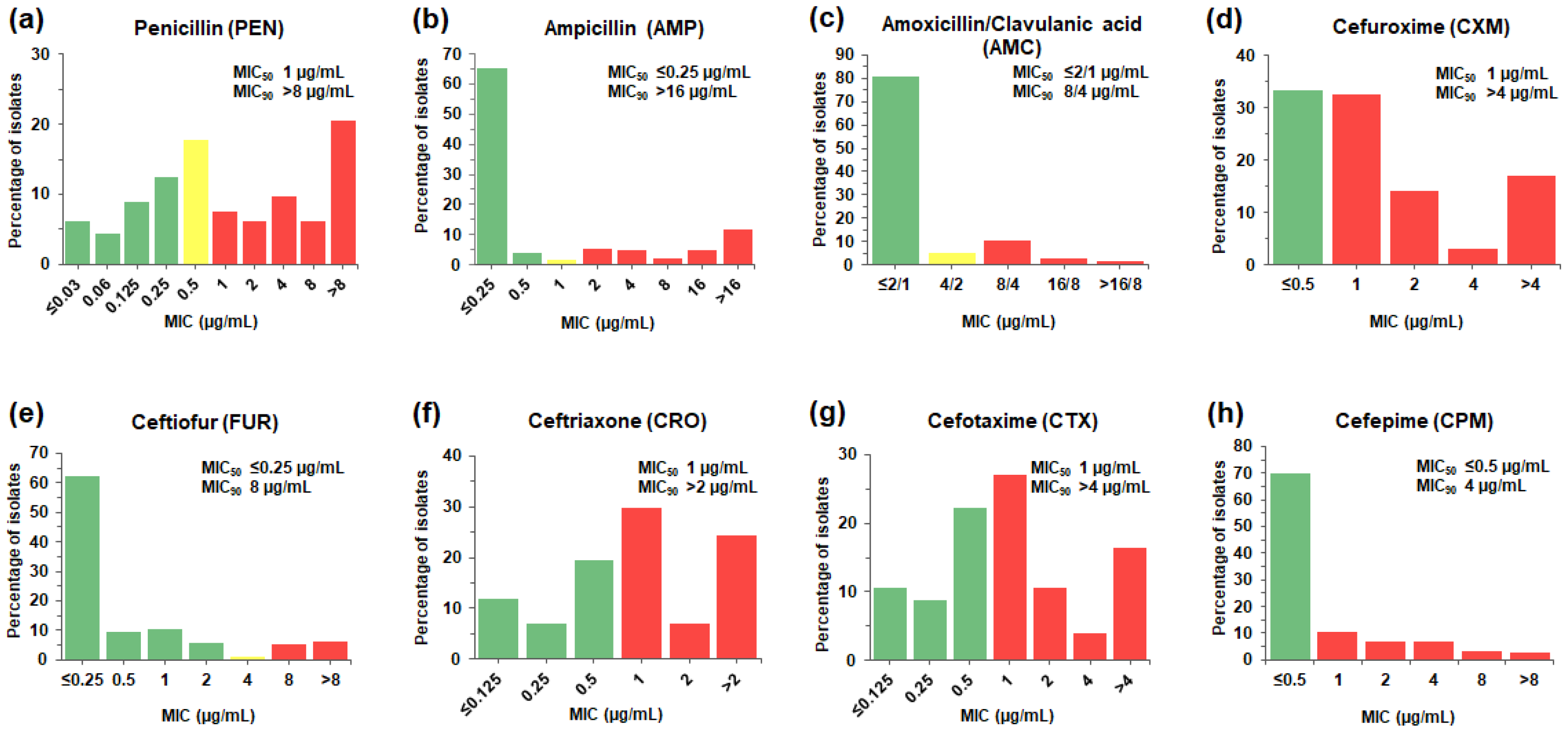
MIC distribution for (a) amoxicillin/clavulanic acid, ampicillin
4.7 (739) · € 29.99 · Auf Lager
Download scientific diagram | MIC distribution for (a) amoxicillin/clavulanic acid, ampicillin, azithromycin and levofloxacin, and (b) cefaclor, cefpodoxime, ceftriaxone, and cefuroxime against H. influenzae. from publication: Results from the Survey of Antibiotic Resistance (SOAR) 2011–13 in Turkey | Objectives: Data are presented from the Survey of Antibiotic Resistance (SOAR) for respiratory tract infection pathogens collected in 2011-13 from Turkey. Methods: MICs were determined using Etest®. Susceptibility was assessed using CLSI, EUCAST and | Turkey, Survey and Surveys and questionnaires | ResearchGate, the professional network for scientists.

MIC distribution for (a) amoxicillin/clavulanic acid, ampicillin

Evaluation and comparison of antibiotic susceptibility profiles of Streptomyces spp. from clinical specimens revealed common and region-dependent resistance patterns
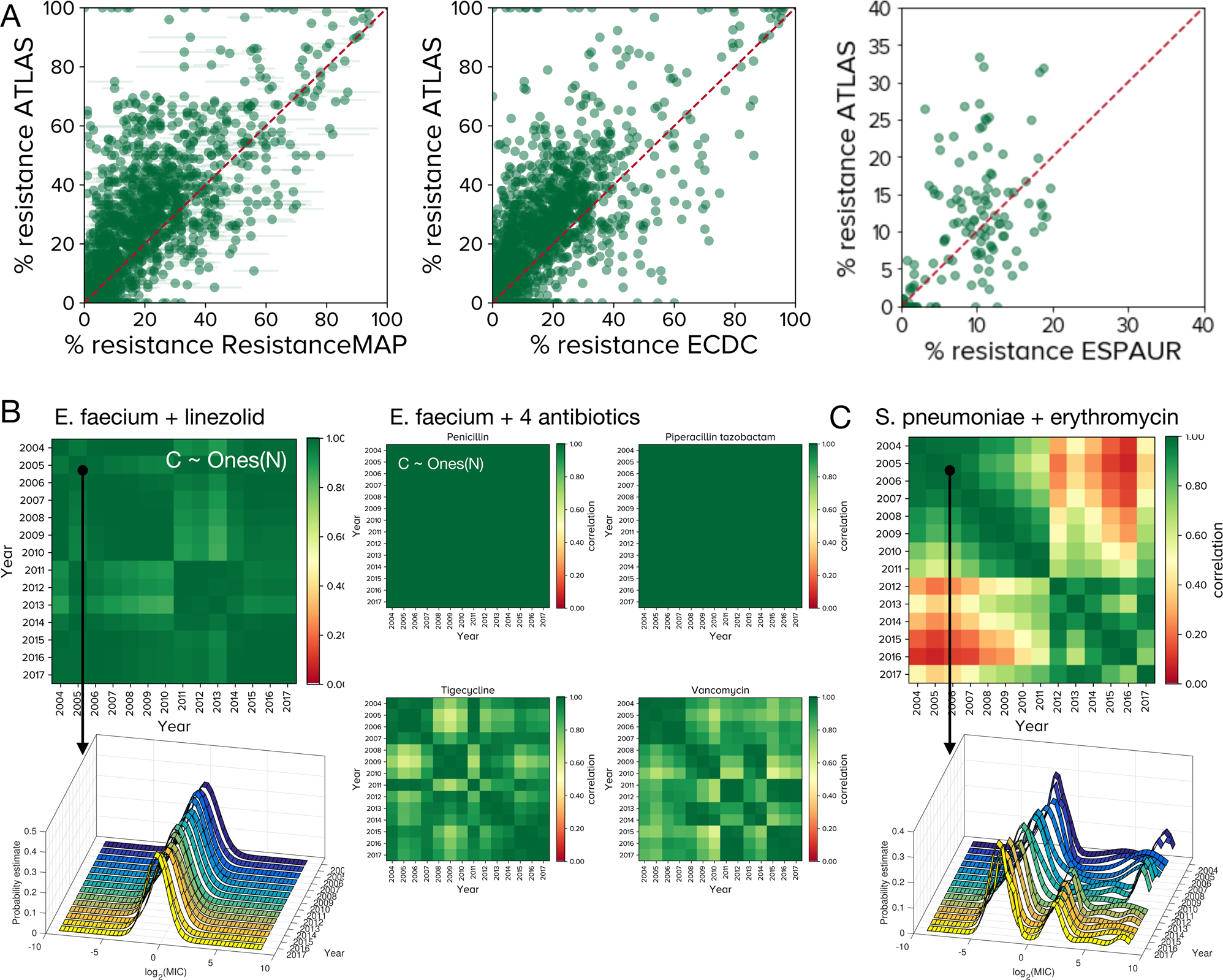
Seeking patterns of antibiotic resistance in ATLAS, an open, raw MIC database with patient metadata

MIC distributions of b-lactam antibiotics for different SCCmec types.

PDF] A combination of mecillinam and amoxicillin/clavulanate can restore susceptibility of high-level TEM-1-producing Escherichia coli to mecillinam

Antibiotic Resistance in Haemophilus influenzae Decreased, except for β-Lactamase-Negative Amoxicillin-Resistant Isolates, in Parallel with Community Antibiotic Consumption in Spain from 1997 to 2007
Antibiotics, Free Full-Text

Is the standard dose of amoxicillin-clavulanic acid sufficient?, BMC Pharmacology and Toxicology

PDF) Is the standard dose of amoxicillin-clavulanic acid sufficient?
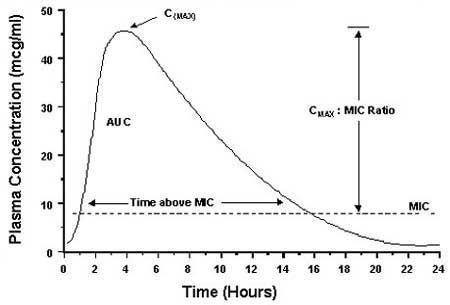
Strategies for Using Antibiotics in Animals - WSAVA2002 - VIN

Ampicillin-Sulbactam and Amoxicillin-Clavulanate Susceptibility Testing of Escherichia coli Isolates with Different β-Lactam Resistance Phenotypes
Frontiers Antimicrobial Resistance Analysis of Clinical Escherichia coli Isolates in Neonatal Ward

Neural network-based predictions of antimicrobial resistance in Salmonella spp. using k-mers counting from whole-genome sequences

Molecular Cut-off Values for Aliarcobacter butzleri Susceptibility Testing

Antibiotics, Free Full-Text