- Startseite
- entfernung membran
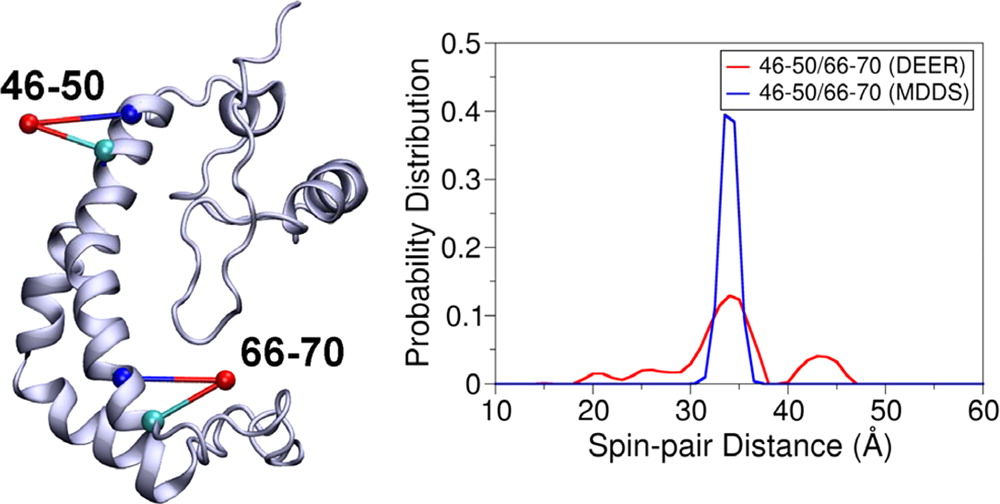
- Simulating the Distance Distribution between Spin-Labels Attached to Proteins
Simulating the Distance Distribution between Spin-Labels Attached to Proteins
4.5 (548) · € 23.00 · Auf Lager
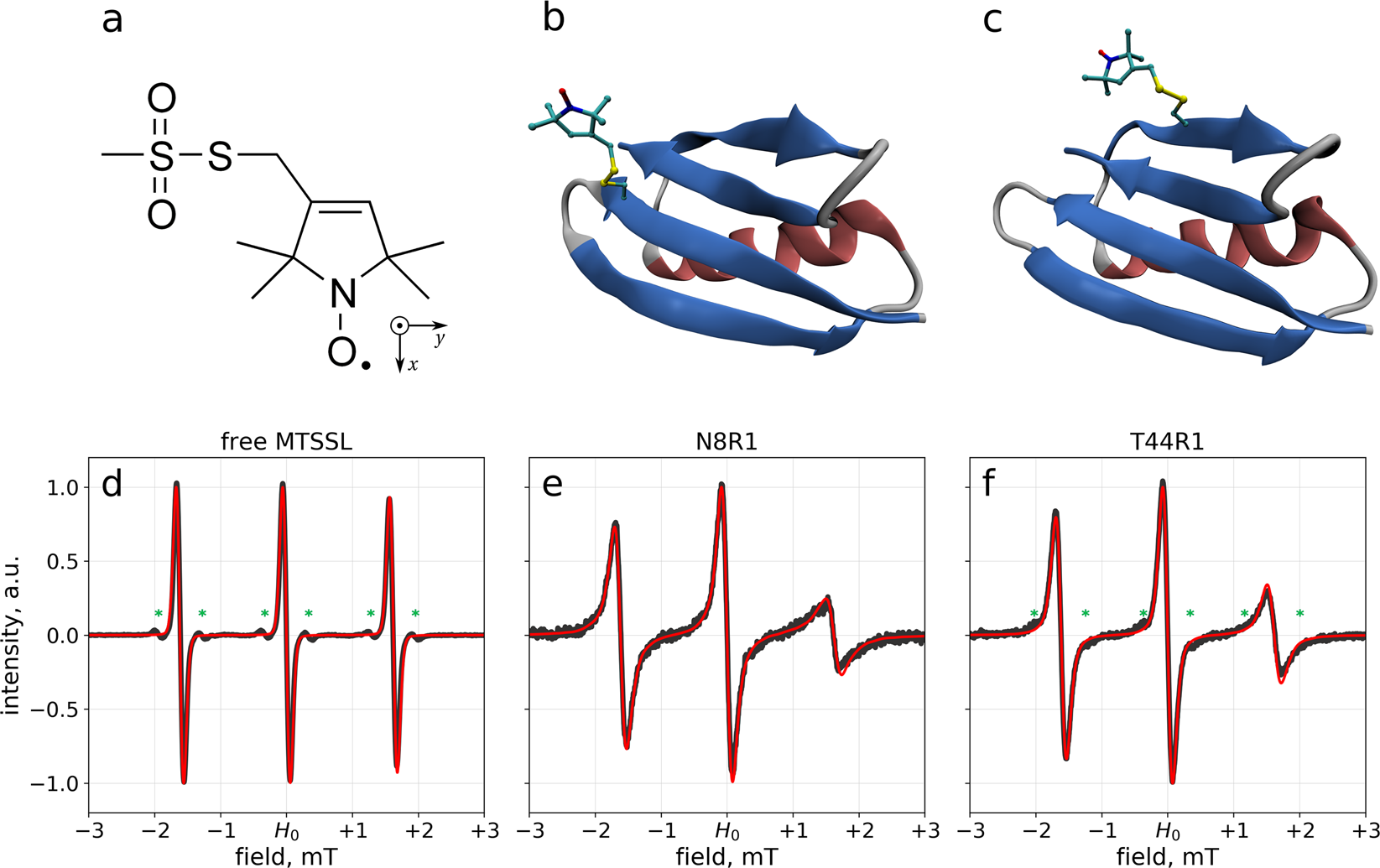
Structural and dynamic origins of ESR lineshapes in spin-labeled GB1 domain: the insights from spin dynamics simulations based on long MD trajectories

Site-directed spin labeling of proteins for distance measurements in vitro and in cells - Organic & Biomolecular Chemistry (RSC Publishing) DOI:10.1039/C6OB00473C
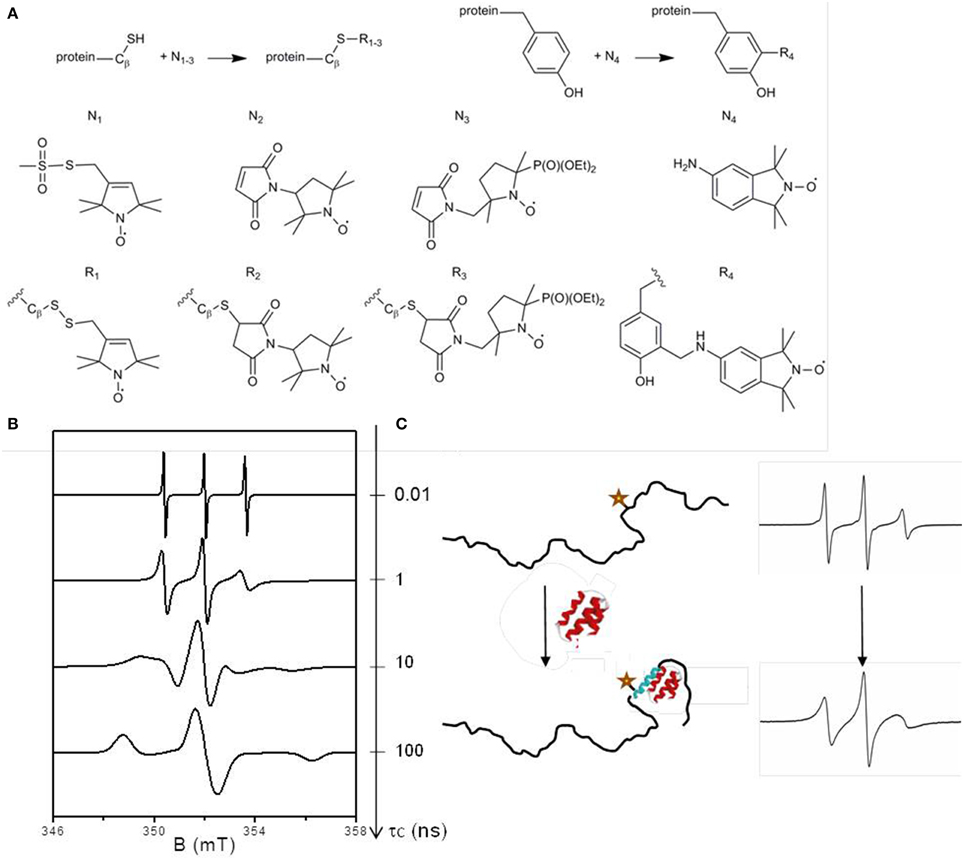
Frontiers Exploring intrinsically disordered proteins using site-directed spin labeling electron paramagnetic resonance spectroscopy
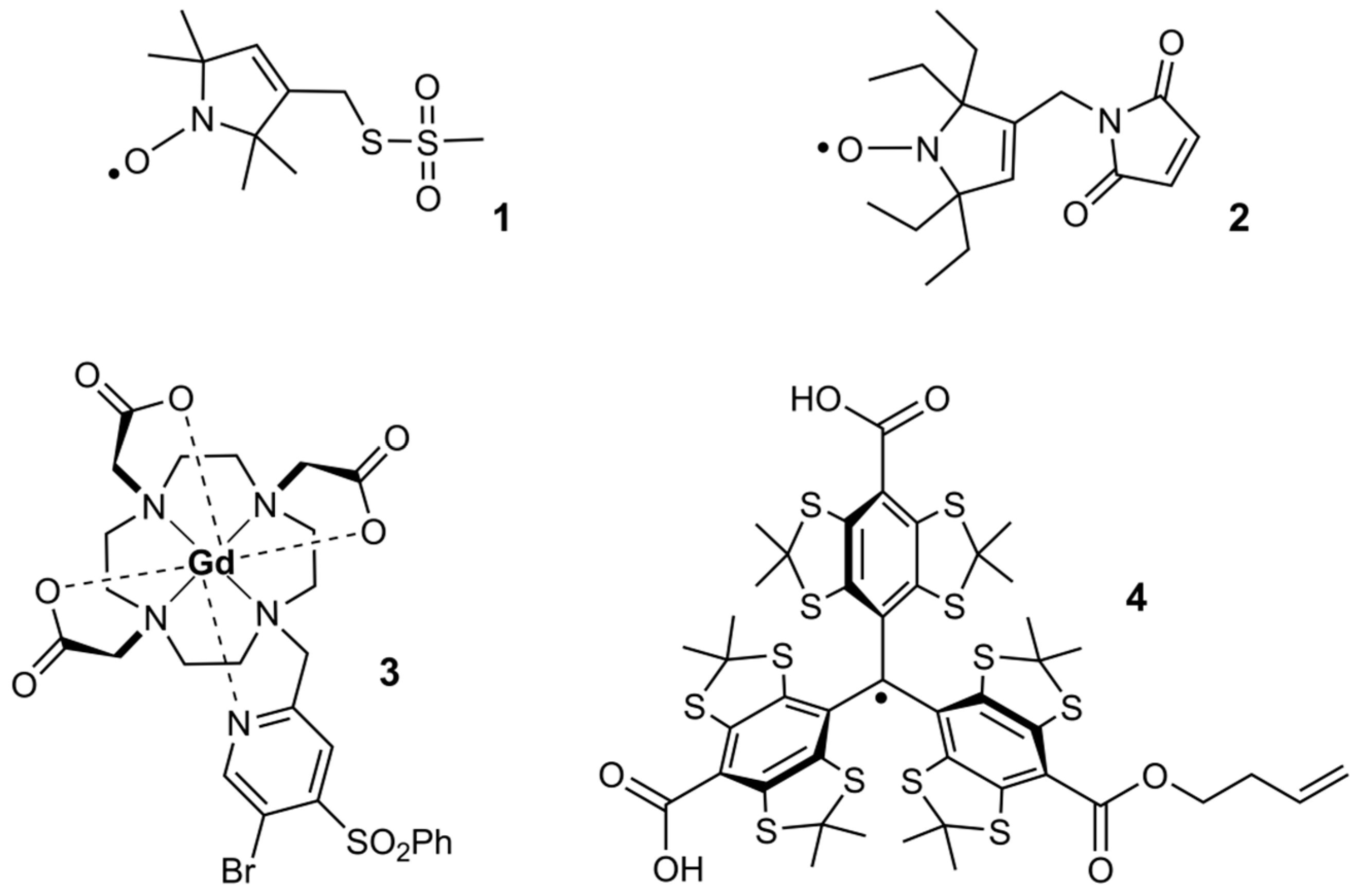
Molecules, Free Full-Text

New limits of sensitivity of site-directed spin labeling electron paramagnetic resonance for membrane proteins - ScienceDirect
Structural Dynamics of Protein Interactions Using Site-Directed Spin Labeling of Cysteines to Measure Distances and Rotational Dynamics with EPR Spectroscopy

Identifying Protein Conformational Dynamics Using Spin‐label ESR - Lai - 2019 - Chemistry – An Asian Journal - Wiley Online Library

Frontiers Characterization of Weak Protein Domain Structure by Spin-Label Distance Distributions

Enzymatic Spin-Labeling of Protein N- and C-Termini for Electron Paramagnetic Resonance Spectroscopy

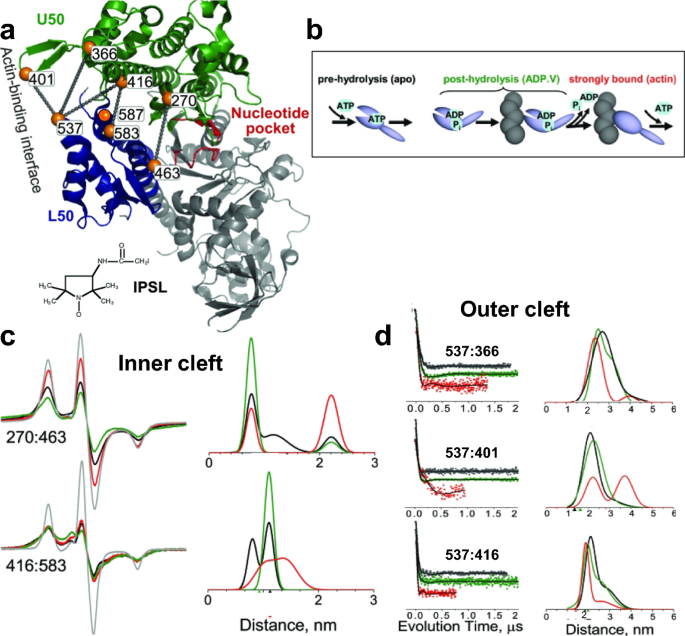
The main simulated distances (black) of PcrA expressed in Å between the